Welcome !
A brief biography
During my academic career, first I examined the conformational changes of DNA molecules induced by local defects on the molecule (bending, kick..) or by changes in global physico-chemical conditions (ionic strength, temperature..). This work aims to probe the physical and mechanical properties of double-stranded DNA molecule which regulating its stiffness and influencing its interaction abilities. To do so, I was performing and combining both experimental measurements, numerical simulation (based on Kinetic Monte Carlo algorithm) and continuously employing and manipulating theoretical descriptions of bio-polymer models at various order of approximation. This interdisciplinary way of investigating and analyzing high-throughput data motivate me to start working at the chromatin scale (all genomic material inside the nucleus).
Secondary, during my post-doctorant's projects, next during my Researcher projects (as projects leader of Computational genome modeling section in CollasLab group), studies focus on investigating the large-scale 3-Dimensional organization of the genome inside cells nucleus. My main project focus on establishing how lamina-associated domains (also called LADs) could modulate mechanical constraints and physical properties of the chromatin fiber ? How these lamina-associated domains could dynamically contribute in regulating genes expressions or transcriptions activities of those or neighborhood chromatin regions. Combine big data analysis (RNASeq, ChIPSeq data) with Monte-Carlo Simulations based on a mesoscopic statistical model of chromatin allow us to examine gene expression pattern over time and 3D genome organization in various context (cancerous, circadian context see Oscillating pattern of DNA-lamins interactions at the NM , cell type specificity..).
Please find additional information on scientific projects at Research page and a list of published scientific articles in the Publication page.
From the DNA molecule to the chromatin fiber inside cell nucleus
Genetic information is encoded on the DNA molecule. The DNA is considere as a bio-polymer : succession of components (each component could be see as "block") sharing similar properties. In human cells, DNA is found inside the nucleus of the cell. At this scale, we talk about chromatin fiber, genome or genomic material. This distinction emphasize that in native condition (meaning inside cell) the DNA molecule is not a naked molecule but a molecular complex composed of DNA, binding and shaping protein. This genomic material is make up by 46 chromosomes (22 chromosomes pairs + 2 sexual chromosomes). Now, imagine you take one of your cell and if you place end by end each chromosomes and stretched them all the way out as much as you can, we would end up with a ~2 meters long "string" of DNA molecule. (If you do that with all your cells, put together, the resulting length would be about twice the diameter of the Solar System).
If this 2 meters long DNA molecule is randomly pack in a non-ordered "ball" (meaning after your cat played with your ball of yarn, all the unfolded string of yarn lying on the floor, is bring all together, aggregate without trying to well organizing it), the resulting sphere would be a ~100 micromicrometer-size object. This 100 micromicrometer-size non-organized spherical genomic material object can not fit into a cell, typically 10 micromicrometer-size for a human cell, either into its tiny nucleus (human squamous epithelial cell with average of ~30μm diameter-size and ~8μm nucleus size; human red blood cell is composed of a nucleus only of size ~8 μm with a thickness of ~2μm at the periphery ~1μm at the center).
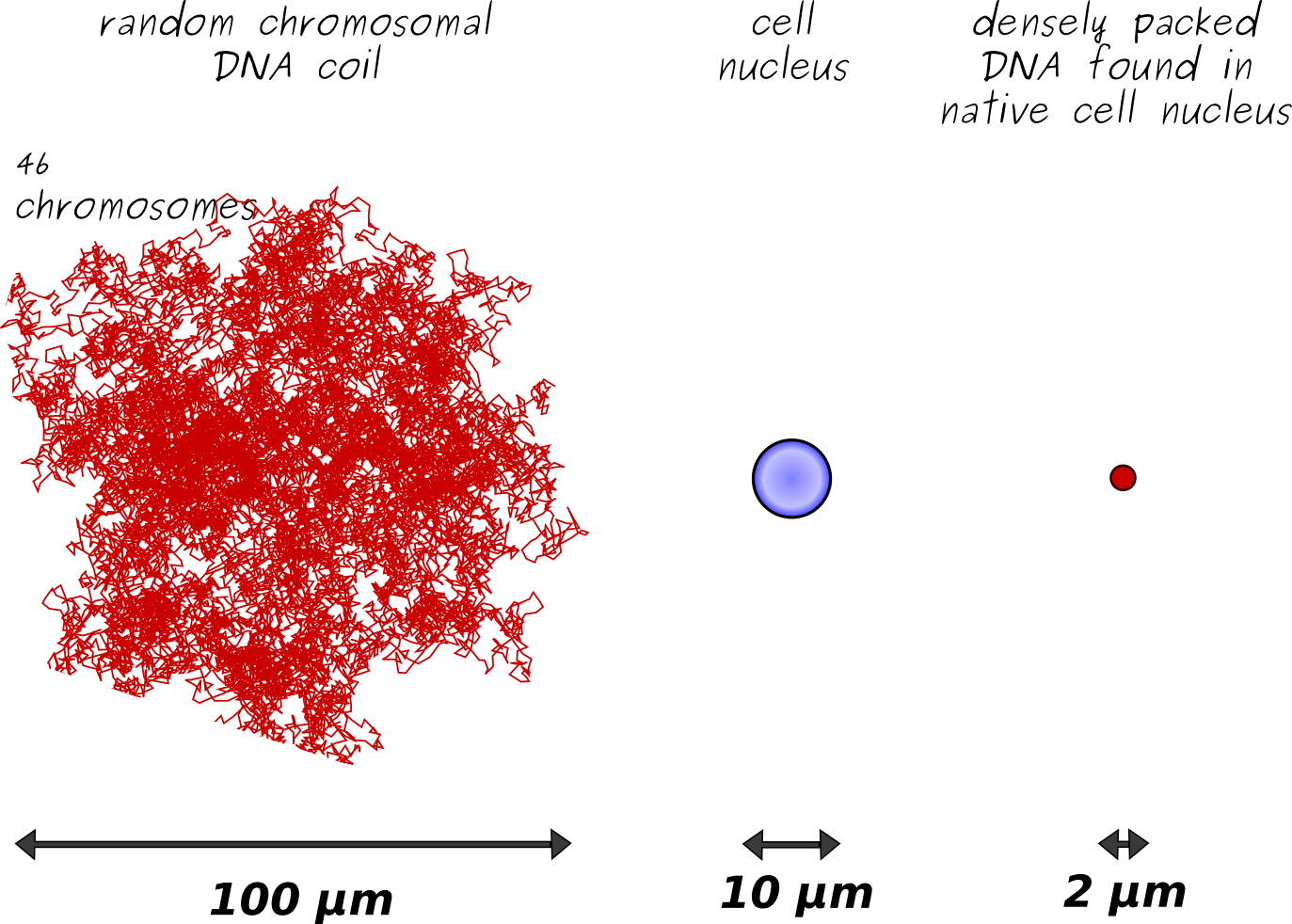
To achieve the necessary compaction of the DNA molecules into the nucleus of the cell, further compaction is needed ! How the DNA, which constitute our genome, undergoes enormous compaction to fit into the tiny space of a cell’s nucleus ? This has been among the major mysteries of cell biology.
To fit the DNA into the nucleus, extra DNA packaging and organization are needed. It has been known for decades that DNA folding is doable thanks to, both, physical properties of the DNA and biological actors interacting with the DNA molecule. The stiffness properties of the DNA (which could be designate under its persistence length or bending modulus characteristics, see project Global ionic strength conditions) play a crucial role on its bendability abilities. In addition, at many level the DNA folding and its 3D organization is mediated by binding proteins and interacting partners which bend, wrap and loop (here, let's only mention the histone proteins. DNA wrap around 8 histone proteins forming a proteins-DNA complex named nucleosome, a fundamental subunit of the chromatin) or anchored the DNA/genomic regions at the periphery of the nucleus (Lamina-associated domains or LADs, are DNA/chromatin region in interaction with lamins proteins, proteins surrounding the inner membrane of the nucleus)
All this folding mechanisms and processes lead to a three-dimensional (3D) organization and architecture of the genomic material in cell nucleus. Last decade have been critical in the huge progress in the study of 3d genome organization and architecture inside the nucleus of eukaryotic cell. The current picture revealed important genome-wide patterns suggesting that chromatin contacts and organization operate at different levels and different orders. Each cell, even in a population of cells, follow similar and main key-pattern of organization.
The double-stranded DNA molecule formed a double helix, composed of two single strands of DNA that run in opposite way to each other and are twist together. Each single strand is made of smaller units called nucleotide, famous A,T,G and C bases, and form a chain. The double-stranded DNA molecule wrapped around histone to form nucleosome. This DNA and histones that are packaged into thin, stringy fibers are call chromatin fiber. Then, this resulting nucleosomal array is organized into high order structure and can exist in various compaction level. Some genomic regions are in either a compact form called heterochromatin region, or less compact form called euchromatin region. Heterochromatin is more compact or closed, gene-poor, transcriptionaly silent regions of the genome (and marked by repressive histone modifications such as H3K9me3 and H3K27me3). Euchromatine in opposition, is more open, gene-rich, transcriptionaly active regions of the genome.
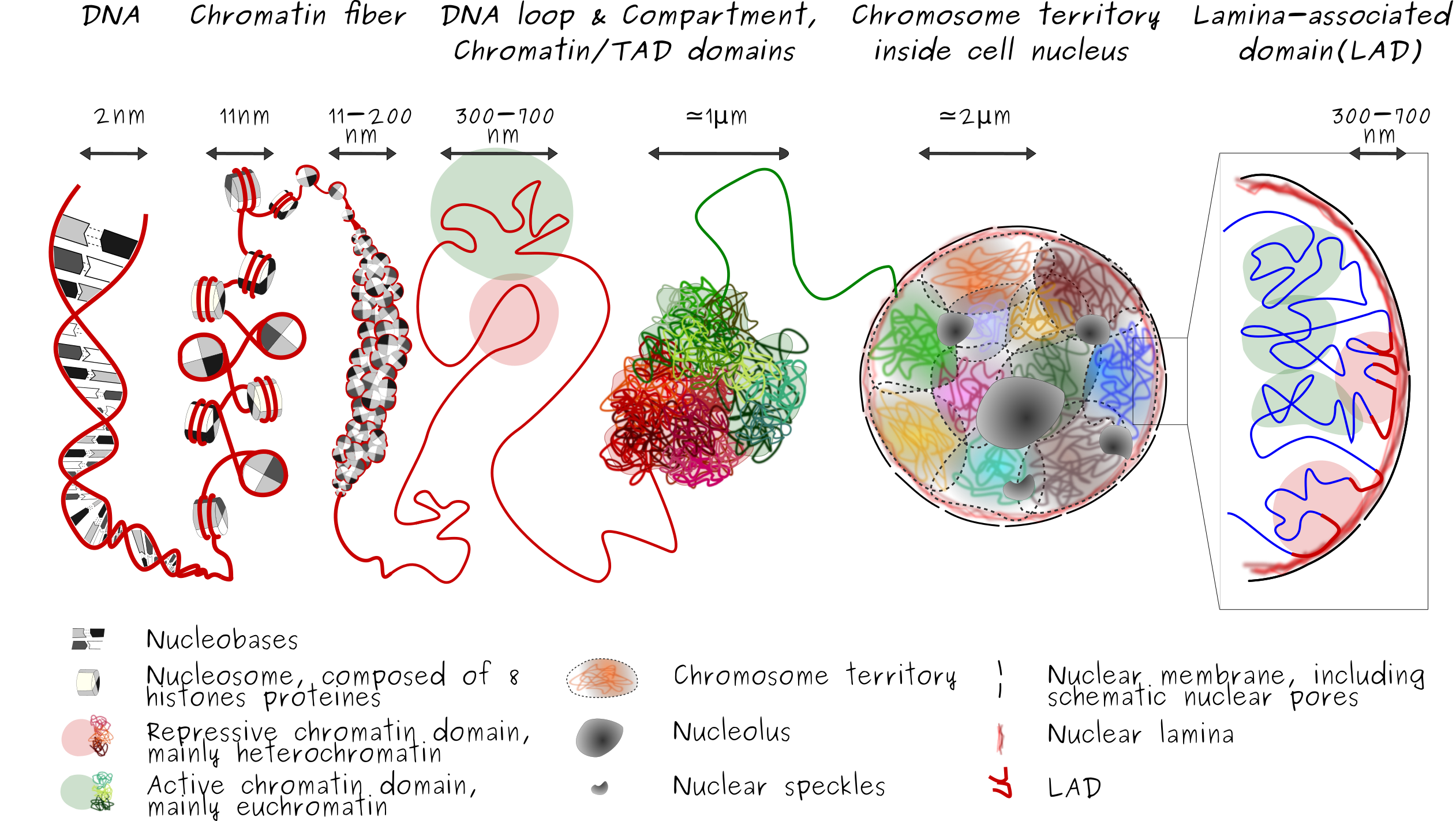
3D conformation of the chromatin is influenced by the specificity of contacts, especially contact with the nuclear lamins proteins at the nuclear lamina (a meshwork of lamins A/C and B poteins) which formed the so lamina-associated-domain or LAD. LADs are typically gene-poor, enriched in heterochromatin and display low gene activity. This layer of organization induce that the genome is also radially organized (i.e., center-to-periphery positioning inside cell nucleus). While lamins A/C, A-type lamins, and B, B-type lamins are localized in the peripheral nuclear lamina, stabilizing heterochromatine in a radial positioning inside the nucleus. A-type lamins also associate with chromatin in the nuclear interior, away from the peripheral nuclear lamina, as for example around the nucleolus. This nucleoplasmic lamin A environment tends to be euchromatic, suggesting distinct roles of lamin A in the regulation of gene expression in peripheral and more central regions of the nucleus, see Differential anchoring strength of lamins protein to DNA at the NM).
Spatial organization and packaging of the genome are important for proper regulation of gene expression and are often altered in disease. Proper positioning of interactions between chromosomes and between chromosomes and the nuclear envelope is crucial for proper regulation of gene expression.
Next-generation sequencing technologies enable mapping these interactions along the genome; however there is little information on how these interactions are mechanistically controlled. Models of 3D chromosome conformation have been proposed, but these have insufficient “physical reality”. In our research project, we propose to study the mechanics driving the 3D arrangement of the genome. I intend to add physical parameters into statistical models of the 3D genome, to better simulate 3D folding. The aim is to probe the impact of these parameters on the system and extract the most significant ranging effects for the 3D organization of chromosomes, see Local modelling of LAD formation dynamics.
